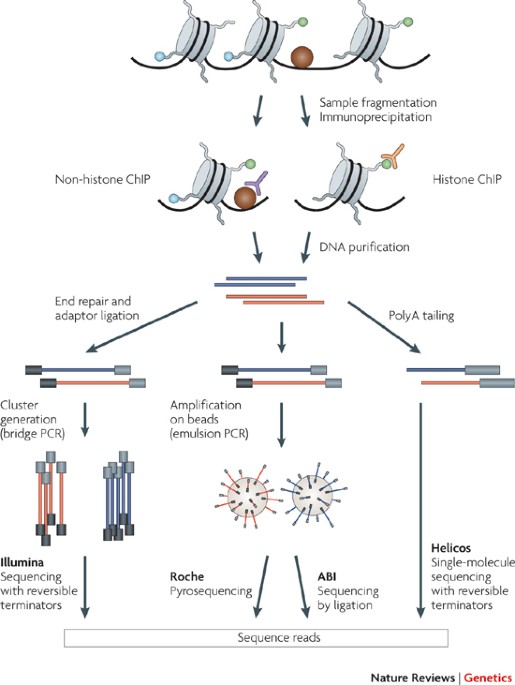
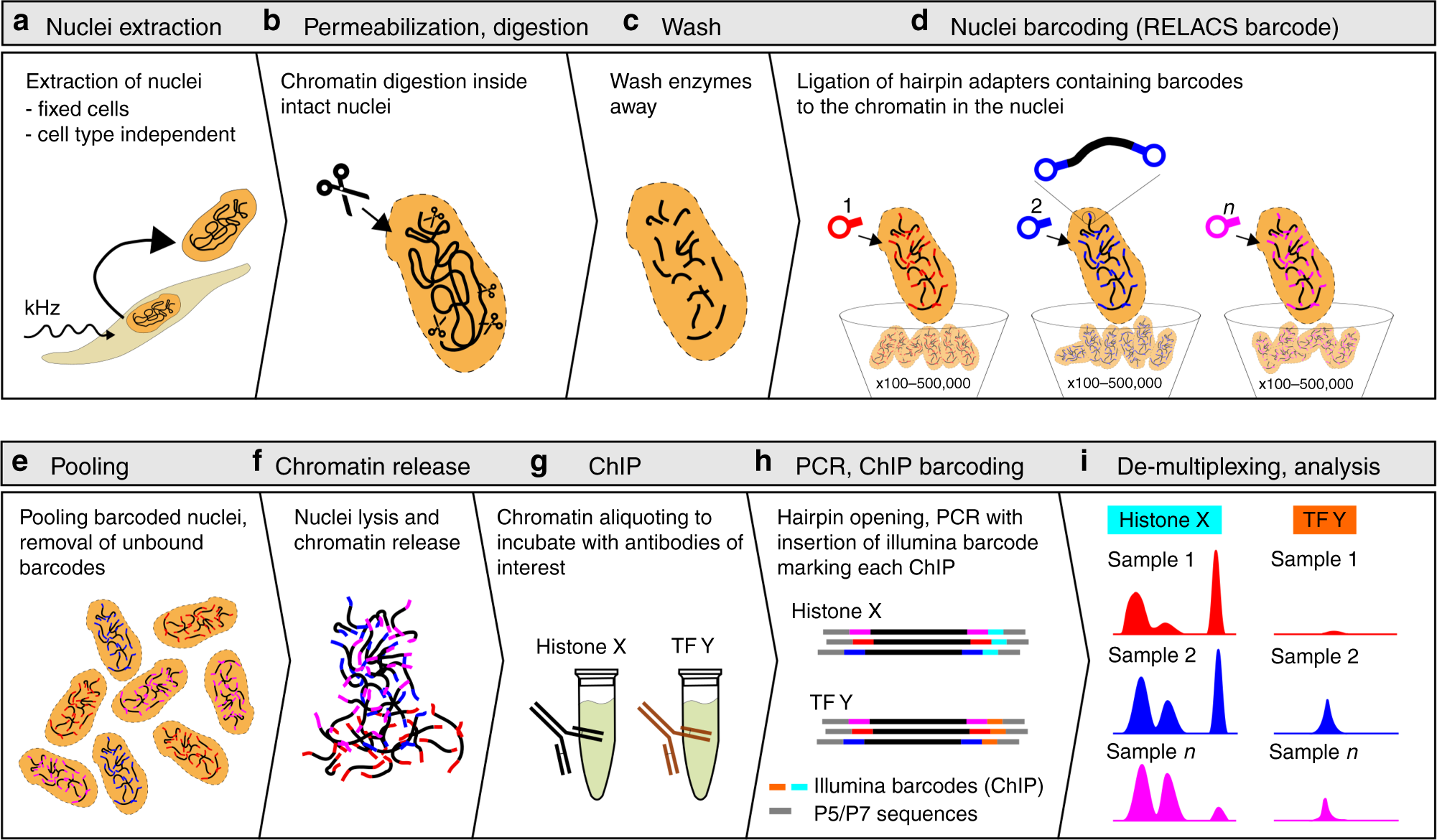
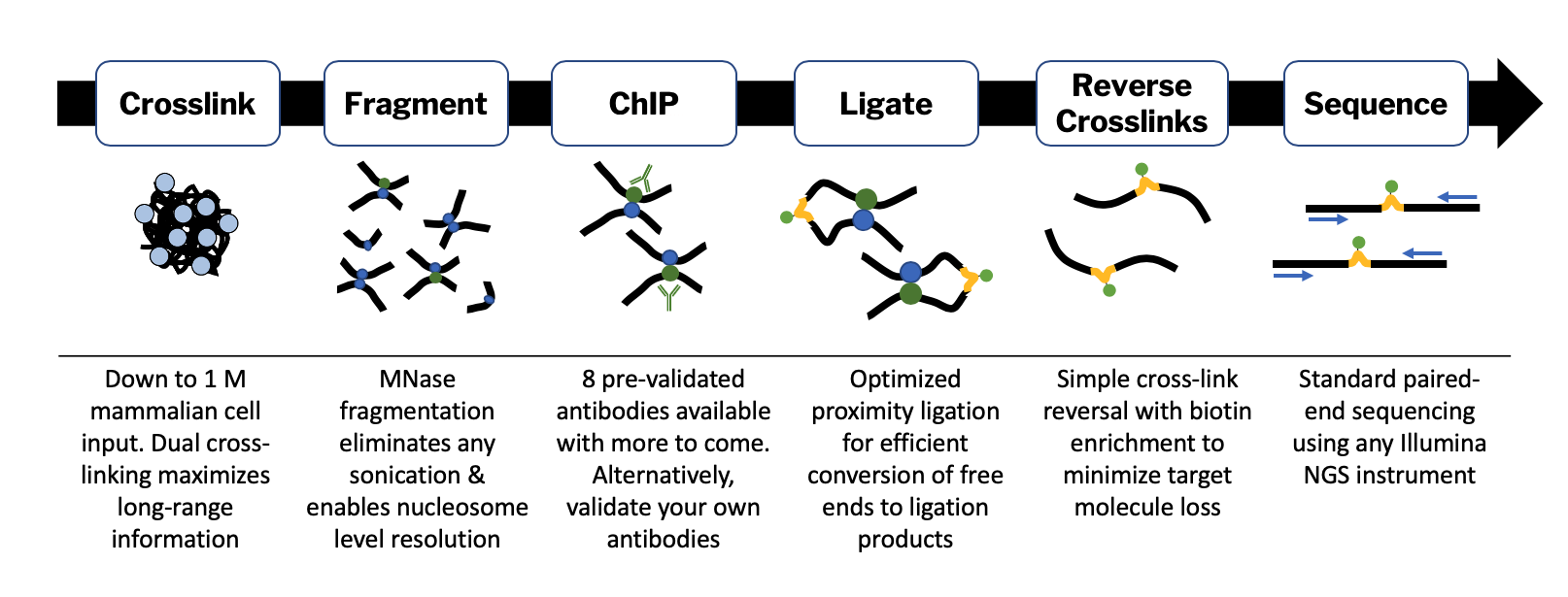
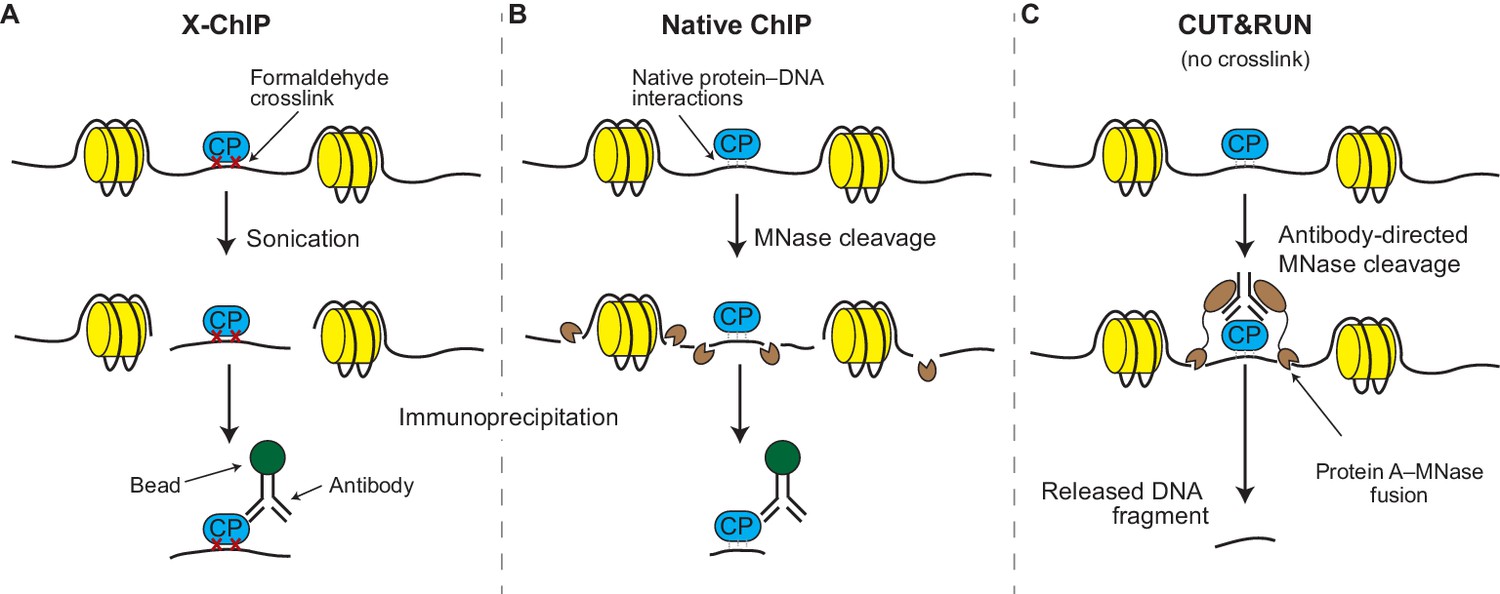
An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations | Nature Communications
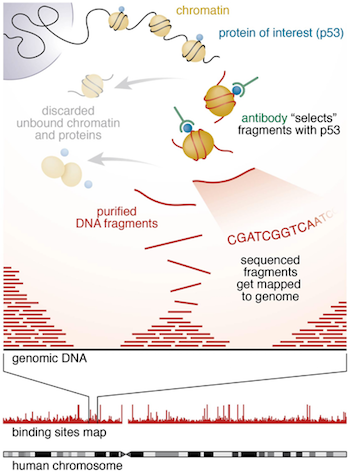
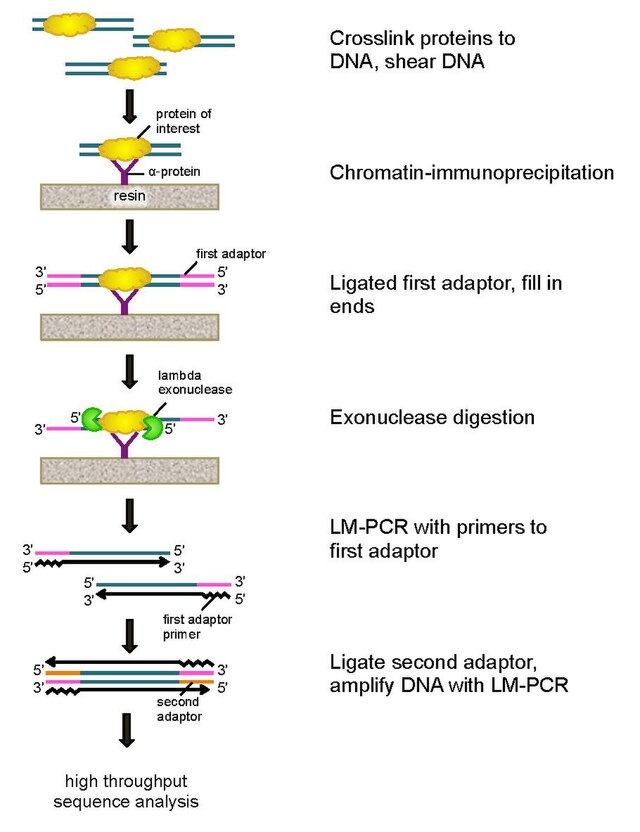
Introduction to ChIP-seq and directory setup | Introduction to ChIP-Seq using high-performance computing
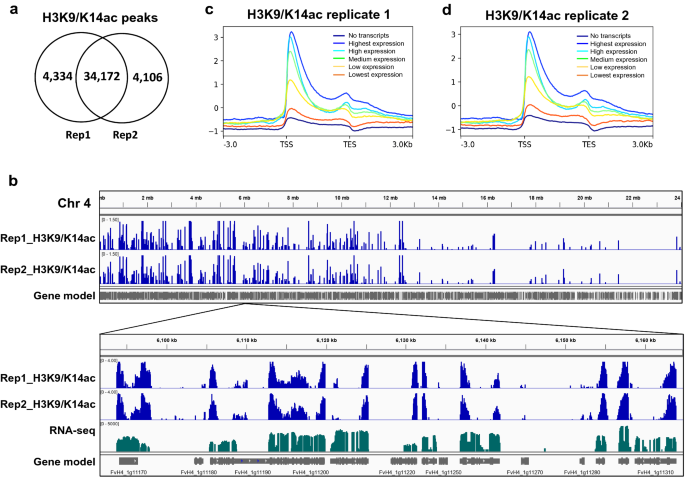
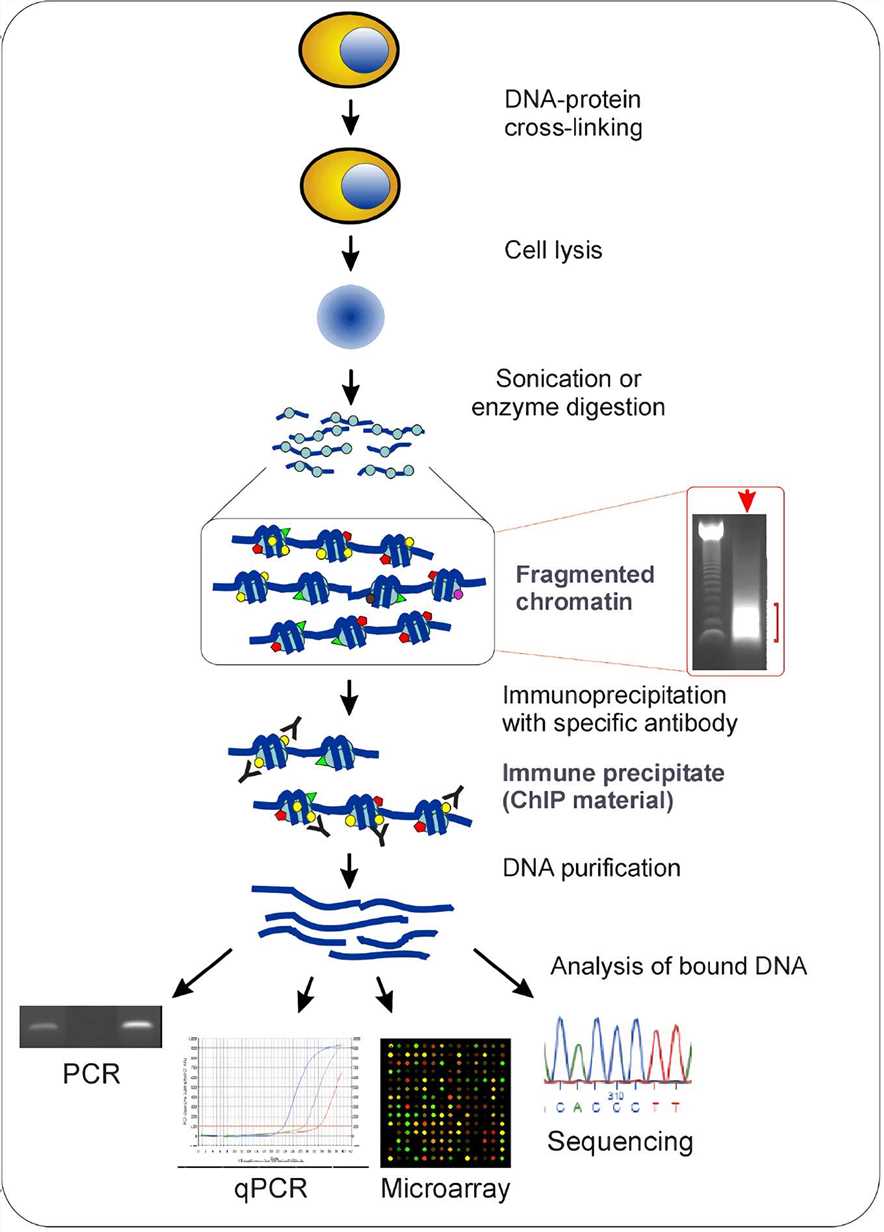
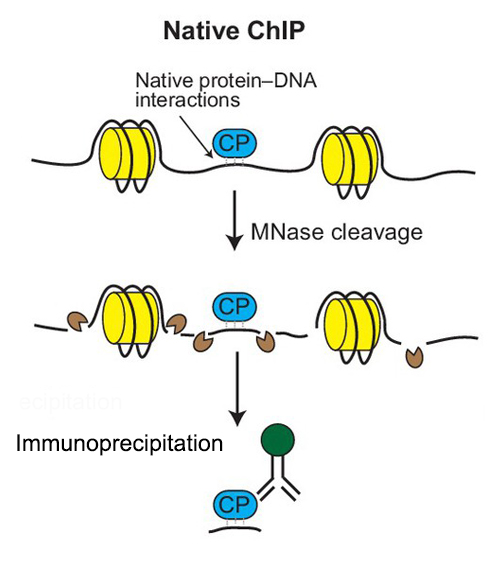
A native chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in strawberry fruits | Plant Methods | Full Text

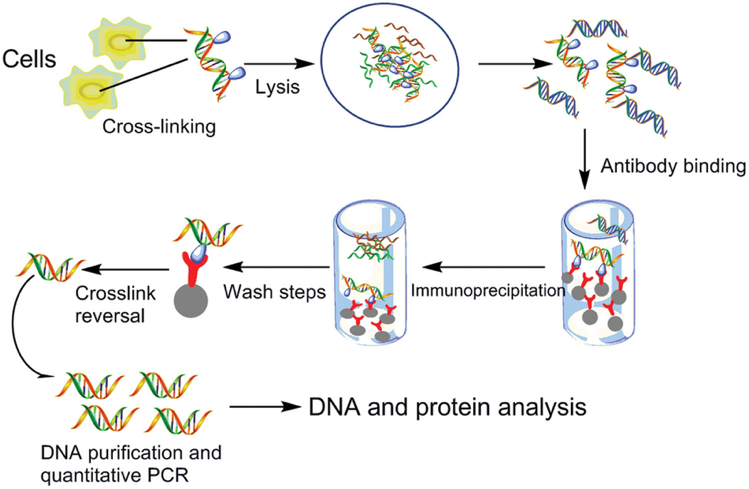
Protocol for fractionation-assisted native ChIP (fanChIP) to capture protein-protein/DNA interactions on chromatin - ScienceDirect

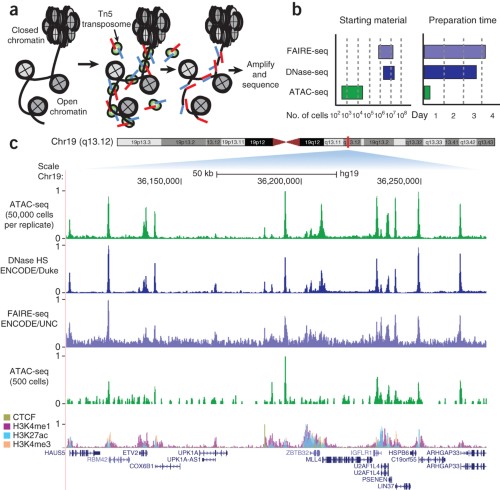
ChIP-seq. a The schematic illustrates conceptual design and key steps... | Download Scientific Diagram
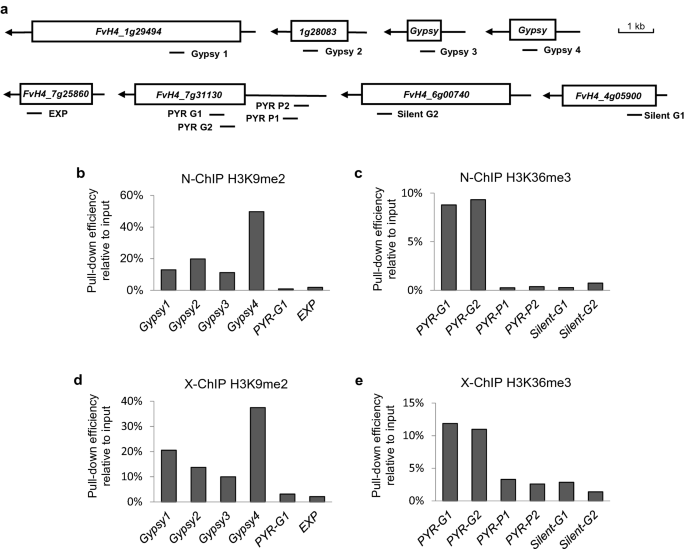
A native chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in strawberry fruits | Plant Methods | Full Text
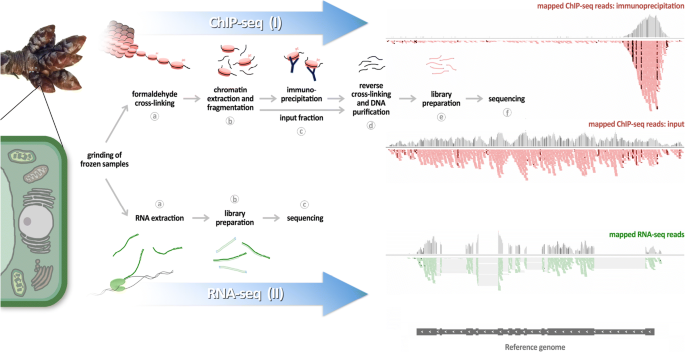
ChIP-seq and RNA-seq for complex and low-abundance tree buds reveal chromatin and expression co-dynamics during sweet cherry bud dormancy | SpringerLink